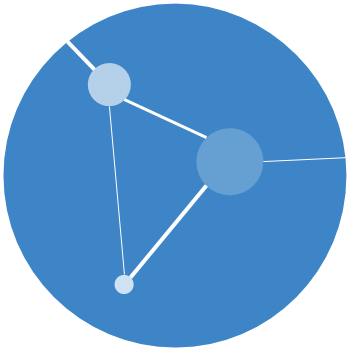
Firstly, what is a homoplasy? It is a trait (for example, a nucleotide at a position in an alignment) that is shared between tips that their immediate common ancestor doesn’t have.
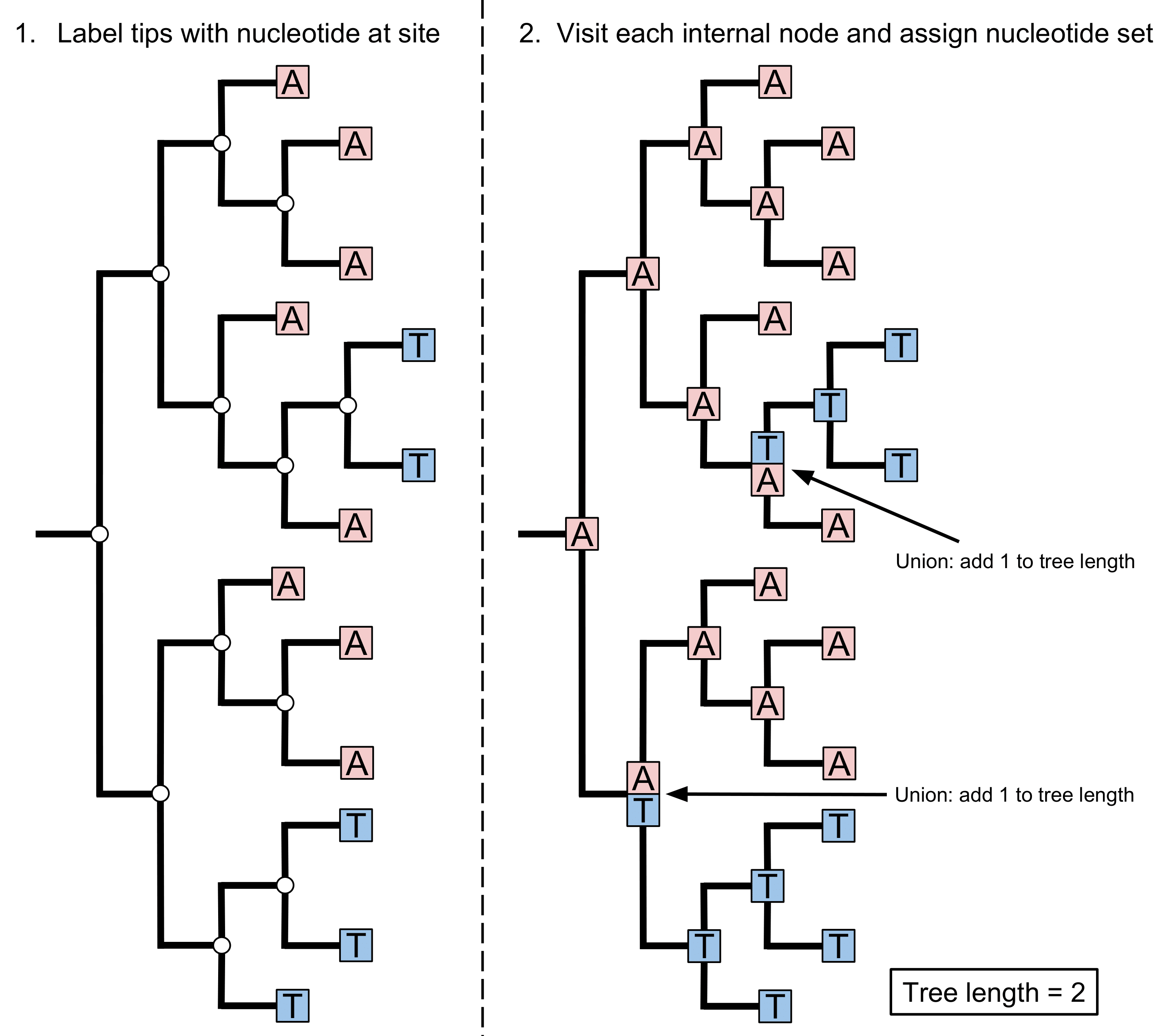
See the left hand side of the figure below, there are tips with a T (thymine) nucleotide that are surrounded by tips with an A (adenine).
Homoplasies are interesting because they make sequences seem more similar than they evolutionarily are - complicating phylogenetic analyses.
Homoplasies can also help us to understand evolution, perhaps there is a benefit to a particular mutation or trait. By identifying homoplasies, we can hone in our search to mutations causing antibiotic resistance, for example.
From the right handside of the figure above, we can see how you can identify homoplasies. Read more about how HomoplasyFinder uses the consistency index to identify here.
Soon after I moved to Dublin, I decided to build HomoplasyFinder. An R package that we can use to identify homoplasies on a phylogeny using a nucleotide alignment.
After a quick look (I should have spent longer! 😳), I couldn’t find a tool so I decided to created my own.
I spent a bit of time trying to figure out how to make my tool so that it could automatically identify homoplasies and came up a pretty good solution (I thought!). So, I created the HomoplasyFinder tool and then spent much longer figuring out whether it worked!
To my surprise, it did work and, happy with my creation, I proceeded to create an R package and write an article. I submitted my article and month later all my hopes were dashed!
The article was rejected… 😭
After a wee bit (a lot) of grumping, I realised that my manuscript was rejected for a very good reason. I hadn’t read about the consistency index - a beautifully simple method for identifying homoplasies that was defined in the 1970s (read the article). Well, I had read about it but I hadn’t understood it! The consistency index is a measure of how consistent each site in a set of genetic sequences is with a given phylogeny. The consistency index can be thought of as a measure of how homoplasious a site is
I took a break from HomoplasyFinder, trying to bury this embarrassing affair.
Then after a month or two, I decided to re-make HomoplasyFinder using the consistency index. I also had a thorough look for tools that could identify homoplasies and I found a few.
Unsurprisingly, my new and improved HomoplasyFinder turned out to be much faster than my previous version and, surprisingly, it was also faster, after some improvements, than the tools I found that did the same thing! (This still worries me a little, maybe I am cheating in some way!)
So, with renewed hope, I updated my manuscript and submitted to a different journal. Following some very helpful reviewers comments it got published!! 🎉
A year on and I am still doing a bit of work on HomoplasyFinder and hopefully I will continue to.
Give it a go and let me know what you think!